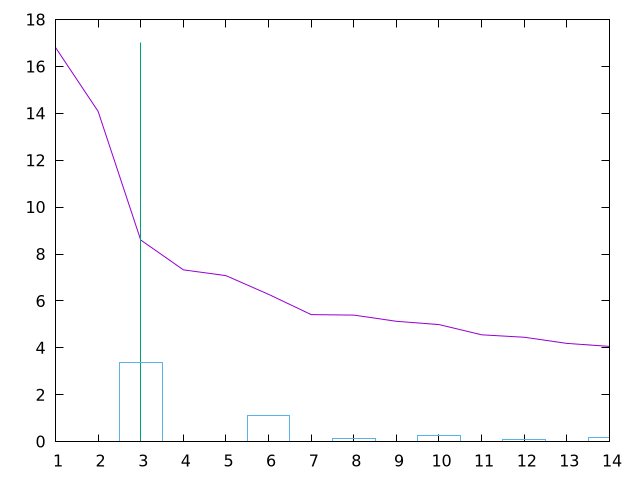
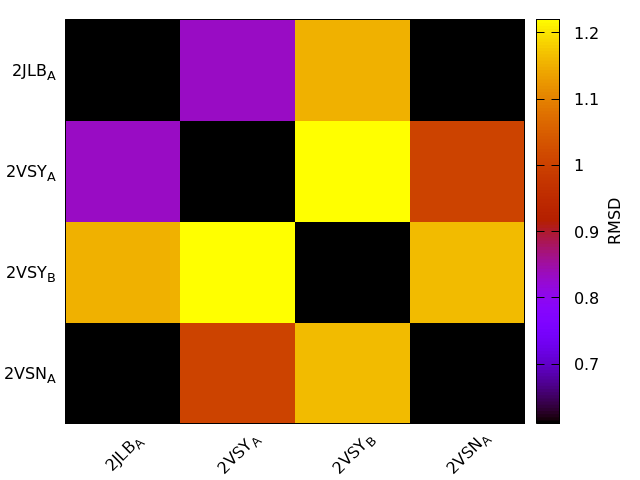
PDBS 5:
2VSN:A 2VSY:A 2VSY:B 2JLB:A
IDENTICAL 2: 2VSN:B 2JLB:B
T-score<0.5 0:
PCA analysis Dimensions 3
PCA analysis Dimensions 4 5
IDENTICAL 2: 2VSN:B 2JLB:B
T-score<0.5 0:
Kpax Structure Alignment
Kpax alignment 5 chains 5 npairs representative 3/0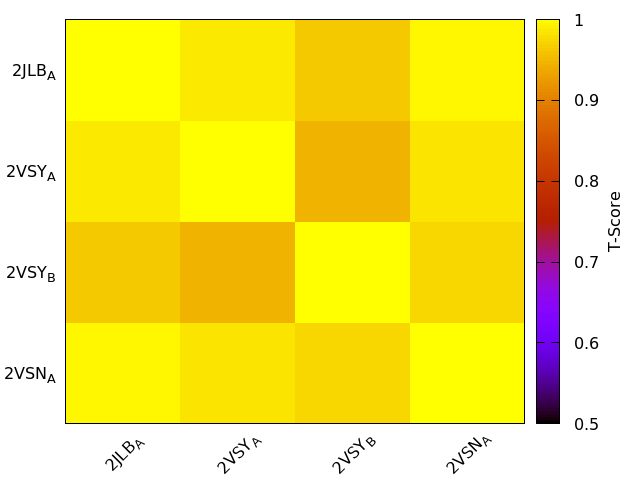 |
 |
 |
Motion 2VSY_A 2VSN_A
Structures
Normal Mode Anaysis



PCA analysis Dimensions 3
PCA analysis Dimensions 4 5




2VSN_A ----AWLMLADAELG-----DTTAGEMAVQR--------PEAVARLGRVRWTQQRHAEAA
2VSY_A QDFVAWLMLADAELGMG---DTTAGEMAVQRGLALHPGHPEAVARLGRVRWTQQRHAEAA
*********** *********** *********************
2VSN_A VLLQQASDAAPEHPGIALWLGHALEDAGQAEAAAAAYTRAHQLLPEEPYITAQLLNWRRR
2VSY_A VLLQQASDAAPEHPGIALWLGHALEDAGQAEAAAAAYTRAHQLLPEEPYITAQLLNWRRR
************************************************************
2VSN_A LCDWRALDVLSAQVRAAVAQGVGAVEPFAFLSEDASAAEQLACARTRAQAIAASVRPLAP
2VSY_A LCDWRALDVLSAQVRAAVAQGVGAVEPFAFLSEDASAAEQLACARTRAQAIAASVRPLAP
************************************************************
2VSN_A TRVRSKGPLRVGFVSNGFGAHPTGLLTVALFEALQRRQPDLQMHLFATSGDDGSTLRTRL
2VSY_A TRVRSKGPLRVGFVSNGFGAHPTGLLTVALFEALQRRQPDLQMHLFATSGDDGSTLRTRL
************************************************************
2VSN_A AQASTLHDVTALGHLATAKHIRHHGIDLLFDLAGWGGGGRPEVFALRPAPVQVNWLAYPG
2VSY_A AQASTLHDVTALGHLATAKHIRHHGIDLLFDLRGWGGGGRPEVFALRPAPVQVNWLAYPG
******************************** ***************************
2VSN_A TSGAPWMDYVLGDAFALPPALEPFYSEHVLRLQGAFQPSDTSRVVAEPPSRTQCGLPEQG
2VSY_A TSGAPWMDYVLGDAFALPPALEPFYSEHVLRLQGAFQPSDTSRVVAEPPSRTQCGLPEQG
************************************************************
2VSN_A VVLCCFNNSYKLNPQSMARMLAVLREVPDSVLWLLSGPGEADARLRAFAHAQGVDAQRLV
2VSY_A VVLCCFNNSYKLNPQSMARMLAVLREVPDSVLWLLSGPGEADARLRAFAHAQGVDAQRLV
************************************************************
2VSN_A FMPKLPHPQYLARYRHADLFLDTHPYNAHTTASDALWTGCPVLTTPGETFAARVAGSLNH
2VSY_A FMPKLPHPQYLARYRHADLFLDTHPYNAHTTASDALWTGCPVLTTPGETFAARVAGSLNH
************************************************************
2VSN_A HLGLDEMNVADDAAFVAKAVALASDPAALTALHARVDVLRRASGVFHMDGFADDFGALLQ
2VSY_A HLGLDEMNVADDAAFVAKAVALASDPAALTALHARVDVLRRASGVFHMDGFADDFGALLQ
************************************************************
2VSN_A ALARRHGWLGI
2VSY_A ALARRHGWLG-
**********
PDB structures